Tools of Recombinant DNA Technology
Tools of Recombinant DNA Technology: Overview
This topic covers concepts, such as Tools of Recombinant DNA Technology, Enzymes Used in Recombinant DNA Technology, Cleaving Enzymes, DNA Ligases, Restriction Enzymes, Palindromic Nucleotide Sequence, Gel Electrophoresis, etc.
Important Questions on Tools of Recombinant DNA Technology
A recombinant DNA is formed when sticky ends of vector DNA and foreign DNA join. The sticky ends are formed by which of the following enzymes:
Agrobacterium mediated genetic transformation described as natured genetic engineering is used to resist plants from:
Which of the following technique is used for separating the fragments of DNA cut by restriction endonucleases?
Biotechnologists refer to which of the following bacteria as a natural genetic engineer of plants.
Which of the following feature of a vector is required to identify the transformed cell?
In recombinant DNA technology, ‘molecular scissors’ are:
An extra-chromosomal, self-replicating part of the cell that has proven to be a boon to biotechnology is:
The genes (a) ori, (b) ampR and (c) rop are associated with which of the following vectors:
Fragments of DNA segments can be separated by:
Restriction endonucleases are used as:
In genetic engineering, which of the following is used?
Does tetracycline resistant gene contain BamH I recognition site?
In four separate experiments (Table), a vector and a fragment was digested with the indicated restriction enzymes. Both, the digested vector and were purified and ligated. represents the ampicillin resistant gene and Ori is the origin of replication. The resultant recombinant vector was transformed into Escherichia coli cells and incubated in a liquid nutrient medium with ampicillin for hours at . Based on the restriction enzymes used for cloning, choose the correct option.
| Experiment No. | Restriction enzymes used |
| 1 | BamHI and SalI |
| 2 | EcoRI and SalI |
| 3 | KpnI and HindIII |
| 4 | KpnI and SalI |
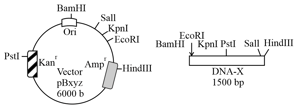
A linear double-stranded DNA was completely digested using either one or two restriction endonucleases ( and ). The sequence of one of the strands is provided. The restriction site for is and for is ( is or and is or ).
Based on this information, the correct option is
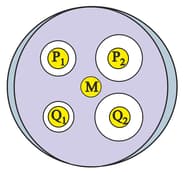
Antibiotics P1, P2 (cell wall synthesis inhibitors), and Q1, Q2 (protein synthesis inhibitors) are equally hydrophilic and moderately toxic to humans. Staphylococcus is known to develop resistance to all these antibiotics. An experimenter tested these antibiotics against a recent pathogenic strain of Bacillus. A high density of Bacillus culture was spread on a nutrient agar plate. Filter paper discs, each soaked in P1, P2, Q1 and Q2 individually (of same concentration), were placed on the bacterial lawn (purple background in the schematic). M is mock control without any antibiotic. Following an overnight incubation, a zone of inhibition is seen around each disc, except for M. Additionally, few individual colonies were observed in zone of inhibition for P2. Based on these observations select the correct statement(s).
Who isolated the antibiotic resistance gene?
Which are the two antibiotic resistance genes present in pBR322?
Antibiotic resistance genes provide antibiotic resistance to their host.
What types of antibiotic resistance genes are present in pBR322?
Define cloning site.
